With Alzheimer’s disease, amyloid peptides combine to form fibrils, in turn forming plaques around neurons. Adobe Stock 421239213
Unexpected Way for Amyloid Peptides to Join Fibrils Explains Time Lag, then Accelerated Growth, in Fibril Formation and Disease Progression
Aggregation of proteins underlies many human disorders, including Alzheimer’s. Teams from the New Jersey Institute of Technology and Princeton University joined forces to study how the amyloid protein aggregates to form plaques, sticky deposits in the brain associated with Alzheimer’s disease. Their simulations, using the second-generation Anton supercomputer developed by D. E. Shaw Research and hosted at PSC, revealed an unexpected mechanism that accelerates aggregation and helps explain the real-world behavior of the molecule. The findings may lead to better treatment and prevention of Alzheimer’s and other human diseases.
WHY IT’S IMPORTANT
When milk curdles, there’s no turning it back. The solids — mostly protein — have clumped together and fallen to the bottom of the carton. And they’re not going to unclump.
Unfortunately, there are proteins and short protein-like molecules called peptides in our bodies that can do the same trick. In the case of milk, it’s bacteria that cause the milk proteins to aggregate. But in disease processes like Alzheimer’s, Parkinson’s, diabetes, and many others, such aggregation can happen without that kind of external jolt. These proteins can self-assemble, in each case causing a critical organ in the body to lose its proper function.
Doctors would like to understand the phenomenon of protein aggregation better. Why do proteins made by our bodies sometimes stick to each other, causing problems? What guides the self assembly process? Can scientists design drugs that prevent it from happening — treating, or even preventing these diseases?
“I think one of the challenges in trying to get to the bottom of these questions … is that the length scales and the time scales that are involved in this process span many, many, orders of magnitude. And so you go from individual peptides to small protofibrils that then grow into these fiber bundles, and so we already have [three or four] orders of magnitude in length scales that in principle one has to account for — and the same goes for time scales.”
— Mikko Haataja, Princeton University
Scientists Cristiano Dias of the New Jersey Institute of Technology and Mikko Haataja of Princeton University joined their teams to study the process of self assembly of a representative amyloid peptide, a process believed to underlie Alzheimer’s disease. To simulate the complicated steps of aggregation, they turned to Anton, a special-purpose supercomputer for molecular dynamics simulation that was designed and constructed by D. E. Shaw Research (DESRES). A second-generation Anton machine is made available without cost by DESRES, and is hosted at PSC with operational funding support by the National Institutes of Health.
HOW PSC HELPED
Amyloid peptides can aggregate on their own, into what are known as fibrils — slender threads of protein. But they don’t do so in a single, simple step. With the version of the peptide present in most humans, plaques of aggregated amyloid fibrils can take many decades to form in the brain. But the interactions between individual peptide molecules take place in the tiniest fraction of a second. Amyloid plaques range from about 50 to 100 micrometers in size — about one three hundredth of an inch. But the chain of amino acids that make up the amyloid peptide is only about 14 nanometers long — over five thousand times smaller.
Simulating the process, then, would span an extraordinary range of times and sizes. Moving from the tiny forces that cause two amyloid peptides to interact to the large structures that aggregation forms would require massive computational power.
Anton at PSC fit the bill for running these complex simulations. This second-generation Anton machine can simulate large numbers of atoms roughly 10 times faster than general-purpose systems. This gives it the ability to simulate molecules for microseconds in a reasonable amount of real time, revealing rare events that can be pivotal for the biomolecules’ functions. In this case, Anton’s superior simulation speed allowed the collaborators to test many assumptions about how the molecules interacted, adjusting the simulated environment until they saw real-world behavior.
“For us, Anton was just critical, because it allowed us to really test [the assumptions behind models for aggregation]. So we said, ‘Oh, let’s run a long simulation — see whether the protein is going to land on the right place [in the fibril].’ Of course it did not. This didn’t happen at the first try, so we had to adjust the conditions for this to happen. So without Anton, it would have taken us several months to just test it, and we would probably have given up the project.”
— Cristiano Dias, New Jersey Institute of Technology
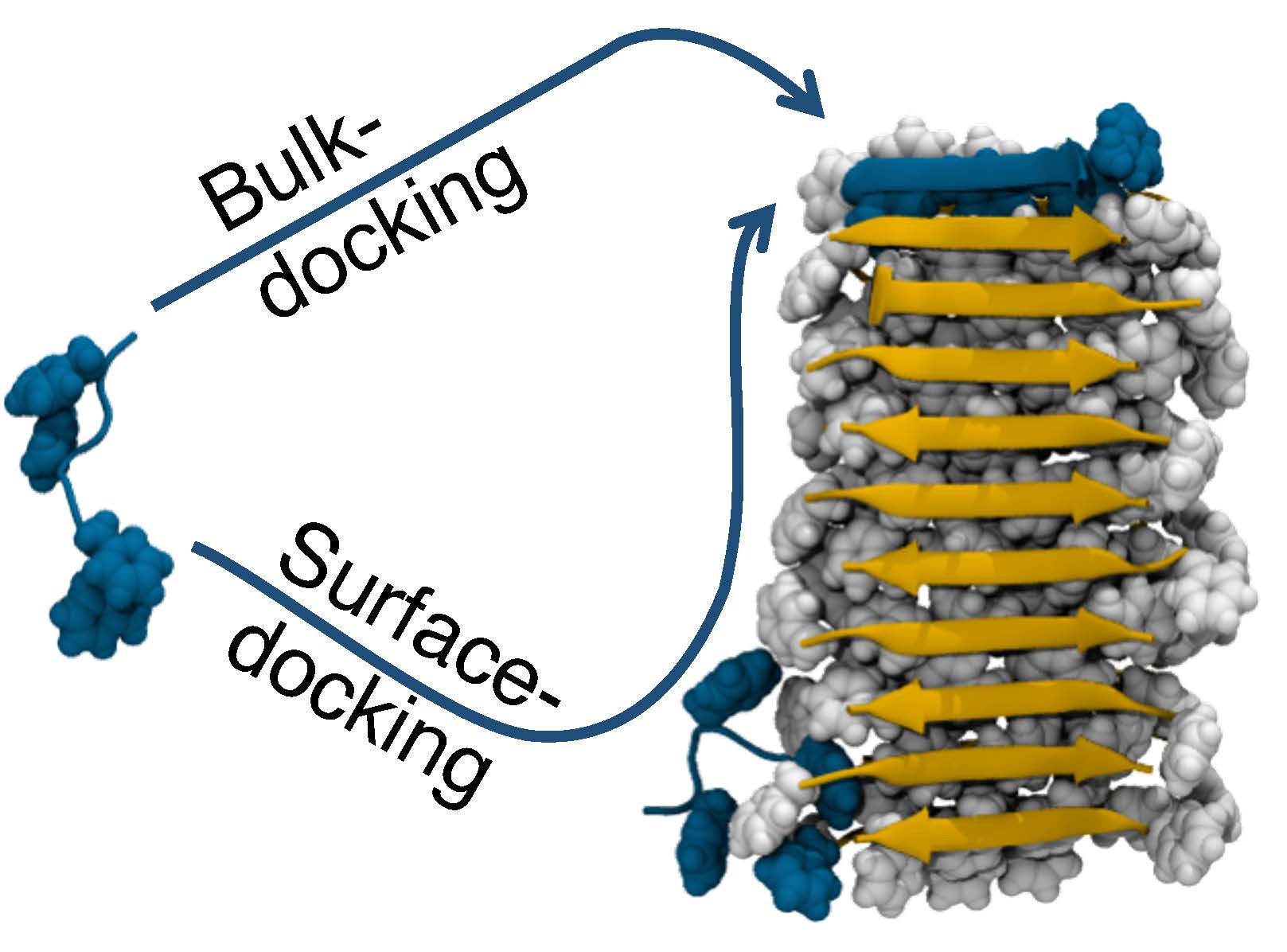
The Anton simulations of amyloid fibril growth revealed that, in addition to sticking to the end of a fibril (Bulk-docking), amyloid peptides (left, blue) could first attach to the length of the fibril (right, gray) and then slide to the end to stick (Surface-docking). This in effect increases the size of the target and makes fibril growth accelerate with time.
The Anton simulations revealed an unexpected behavior by the amyloid peptides. Scientists had known that fibrils grew by new peptides attaching to the end of the fibril. What happened in addition in the simulations, though, was that the amyloid peptides would first stick to the side of the fibril, then slide to the end to attach. In effect, this gave the peptides a much larger target to aim for, explaining how the initial, slow growth of the fibril (when it was short, and so didn’t offer a large target) after a time lag gave way to more explosive growth (once the fibril was longer and easier for the amyloids to “find”).
The discovery challenges previous theoretical “dock-and-lock” models for how amyloid peptides attach to form fibrils and then plaques. The scientists used their new discoveries to create a new model for amyloid growth, which both explains the previous data better and predicts new behaviors by the amyloid peptides and fibrils that can be tested in the laboratory. Ultimately, the scientists hope their work will lead to therapies to treat or prevent Alzheimer’s and many other human disorders. The team, including former PhD students Ruoyao Zhang and Sharareh Jalali, reported their results in the U.S. National Academy of Science’s journal PNAS Nexus in February 2024.
