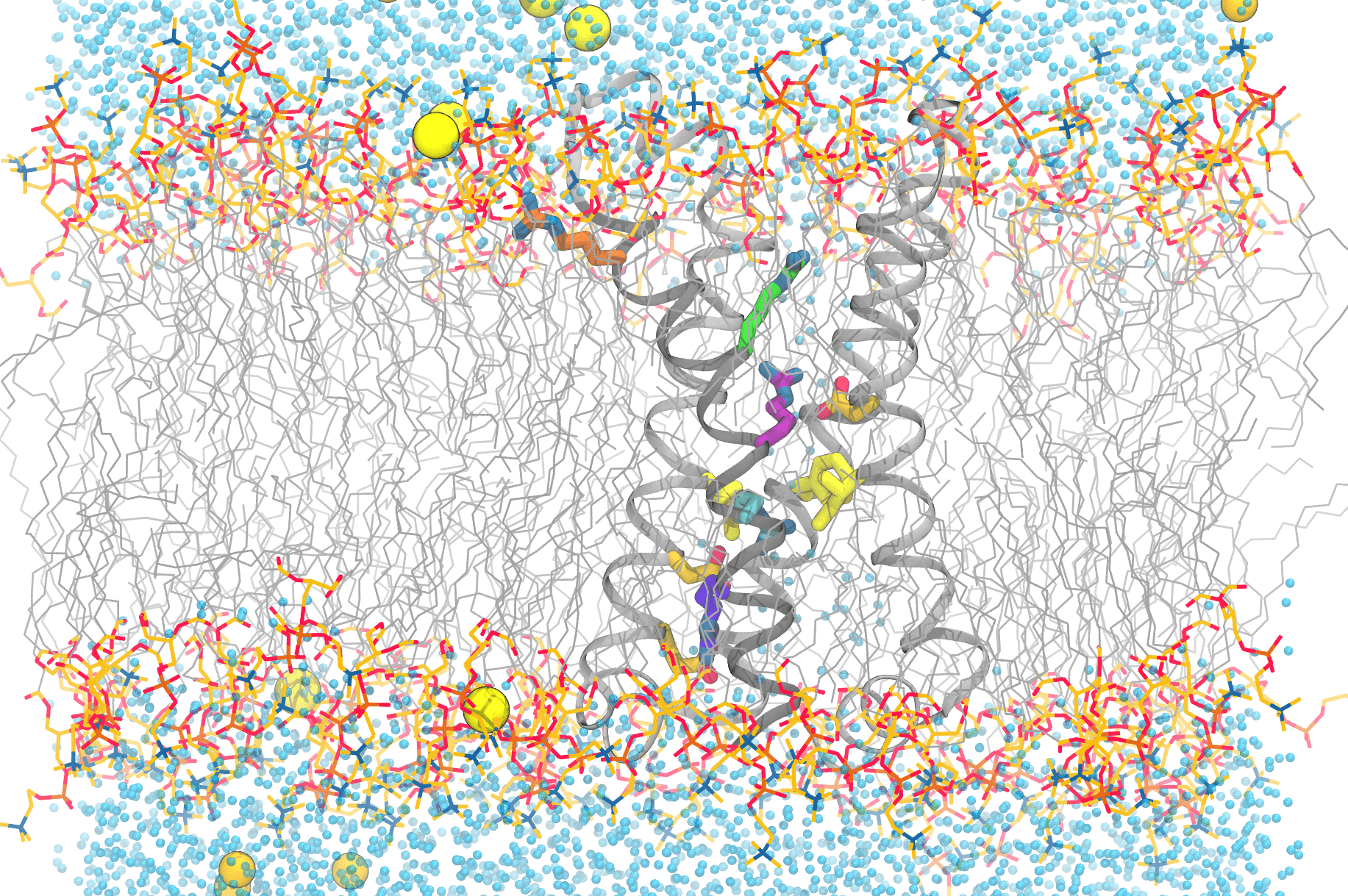
This image shows the complexity of simulating the functional movements of the voltage-sensitive phosphatase’s motions. Anton 2 enables scientists to reproduce the movements of the protein’s components, as well as the lipids (grey, with the headgroups colored), water molecules (blue dots), and ions (yellow/orange spheres) that are essential for a faithful reproduction of the protein in the computer. Credit: Spencer Guo.
University of Chicago Simulations Offer Clues to Behavior of Nerve Cell Channels Underlying Human Diseases
Malfunction of the proteins that sense voltage changes in our nerve cells underlies a number of human diseases throughout the body. A University of Chicago team used an Anton 2 supercomputer developed by D. E. Shaw Research and hosted at PSC to simulate a voltage-sensing protein from a primitive animal to learn how the sensor part of the protein behaves. Their simulations uncovered an unexpected series of motions that hadn’t been predicted by earlier work. The findings gave scientists a new perspective on the complexity of these proteins that may someday help direct design of drugs for human conditions caused by their dysfunctions.
WHY IT’S IMPORTANT
Every movement, thought, and impression we have is a complicated dance of electrical charges running through our brains. Our nerve cells produce electrical signals by first building an imbalance of electrically charged ions (mainly sodium and potassium) across their cell membranes. When a nerve cell receives a signal to fire, it does so by allowing that imbalance to collapse. Tiny channel proteins open up, allowing the ions to flow. As the channels open up, the change in voltage caused by the ion flow causes channels farther down the cell’s long axon to fire up as well. This starts a chain reaction of voltage change down the axon, toward the nerve or muscle cell that receives the signal next. This is called an action potential.
Problems with these voltage-gated ion channels underlie a number of human diseases. In the brain, they can cause epilepsy, ataxia (problems with walking), migraine, deafness, and night blindness. In the electrically sensitive heart muscles, they can cause irregular heartbeats. They can also cause kidney disease and many other conditions.
Doctors would like to understand better how voltage-sensitive channels work. That’s not exactly easy, as they contain two complex parts: the domain that senses voltage changes, and the channel domain itself. But a series of proteins in a number of species have a voltage-sensing domain attached to a simple enzyme protein rather than a complicated channel. These voltage-sensitive phosphatases offer a way of focusing only on the voltage-sensing part.
“When this protein was discovered in other organisms, [scientists] realized [it] has features that are close to those of the channels in our human body, but that is also attached to a different biochemical module, if you will. It’s interesting because in this particular protein you can study the part that’s similar to the human protein, this voltage-sensing module, in an isolated setting.” — Spencer Guo, University of Chicago
Spencer Guo, a graduate student working with Aaron Dinner and Benoit Roux at the University of Chicago, wanted to get a better idea of how the voltage-sensitive domain of the phosphatase works in the sea squirt Ciona intestinalis. This species is a primitive worm-like chordate that is more closely related to us than to actual worms. The experimental structure of this protein was previously determined by Eduardo Perozo’s laboratory. Rong Shen, a staff scientist also involved in the present study, initiated computational studies of it. By simulating how this protein reacts to changes in voltage across a cell membrane, Guo and his teammates could glean clues as to how the more complex voltage-sensitive channels work in humans.
To carry out this challenging series of simulations, the team turned to Anton, a special-purpose supercomputer for molecular dynamics simulation that was designed and constructed by D. E. Shaw Research (DESRES); a second-generation Anton machine is made available without cost by DESRES, and is hosted at PSC with operational funding support by the National Institutes of Health.
HOW PSC HELPED
While the phosphatase Guo was studying would be simpler to simulate in the computer than the channel proteins, the task still wouldn’t be easy. The voltage sensor consists of what for all the world looks like a screw sitting in a socket that itself sits in the cell membrane. X-ray pictures of the motionless protein suggested that the screw might, in response to voltage changes, partly “unscrew” itself, with that motion relayed to the enzyme part of the protein to activate it.
Crucial to visualizing the motion of this protein in action was the Anton supercomputer at PSC, which has the ability to carry out simulations of biomolecules orders of magnitude more quickly than general-purpose supercomputers.
“Most people, when they run simulations, and [particularly on] Anton 2, tend to run very long single or a few simulations … But for our methods, it can be actually more advantageous to run relatively short simulations, and then we statistically combine them … In our case, we still had, I think, something like 400 total microseconds. We ran [about] one microsecond simulation at a time on Anton 2, and that took about 45 minutes to an hour. If you were to do it on a GPU cluster, or the cluster that we have [at the university], that would take about three days at least.” — Spencer Guo, University of Chicago
Guo would need to use Anton a little differently than most scientists. Instead of running very long simulations, which is a unique strength of Anton, he’d need to carry out many short simulations, because the motions of the sensor could be very quick, but could also vary with each repetition because of random movements. By carrying out many short simulations, Guo could sample many examples of the different key motions contributing to activation and characterize their importance statistically — in other words, they could see what the average motion was and how much it differed when repeated.
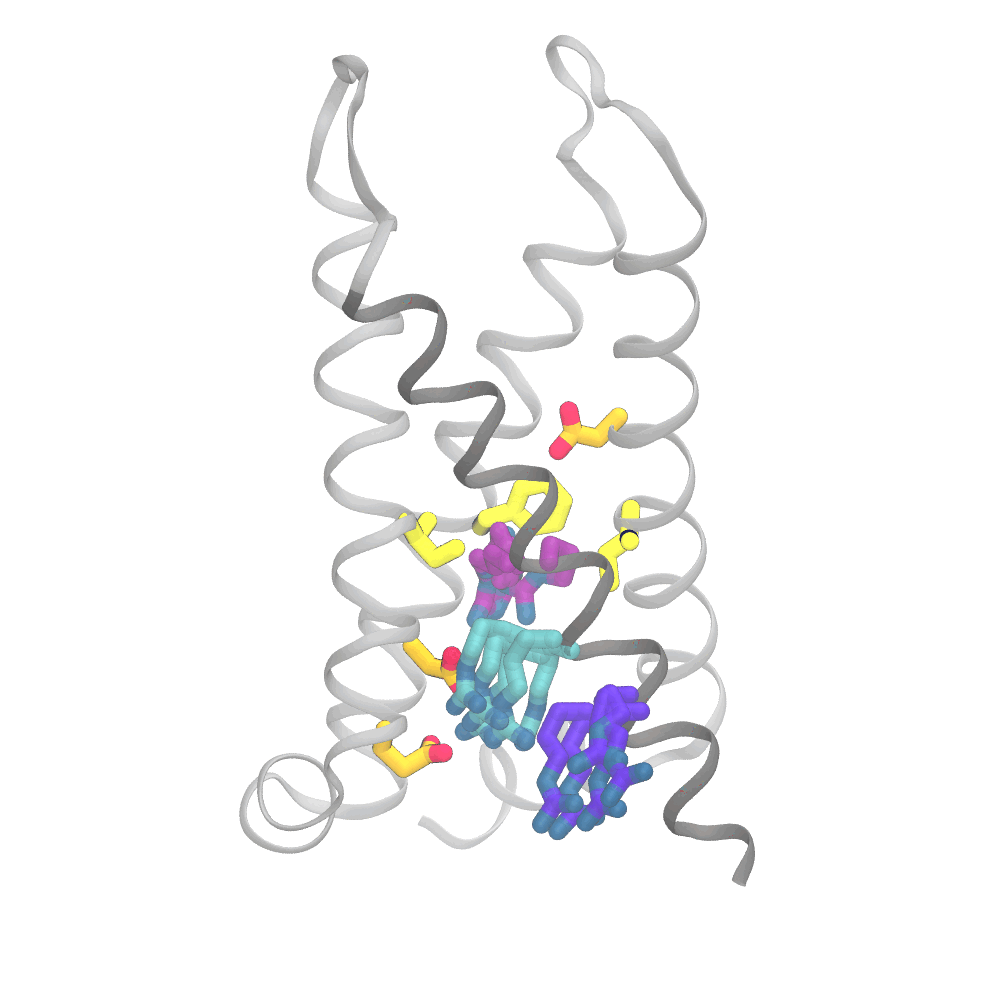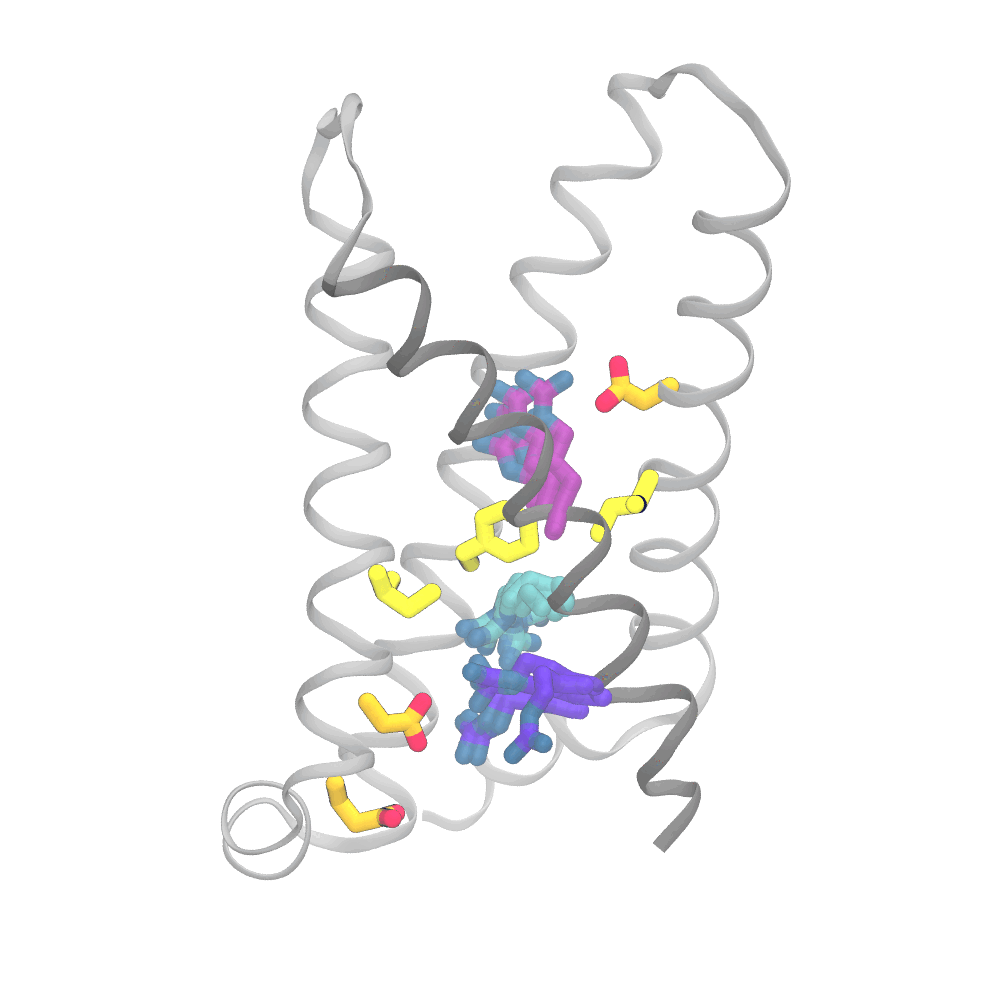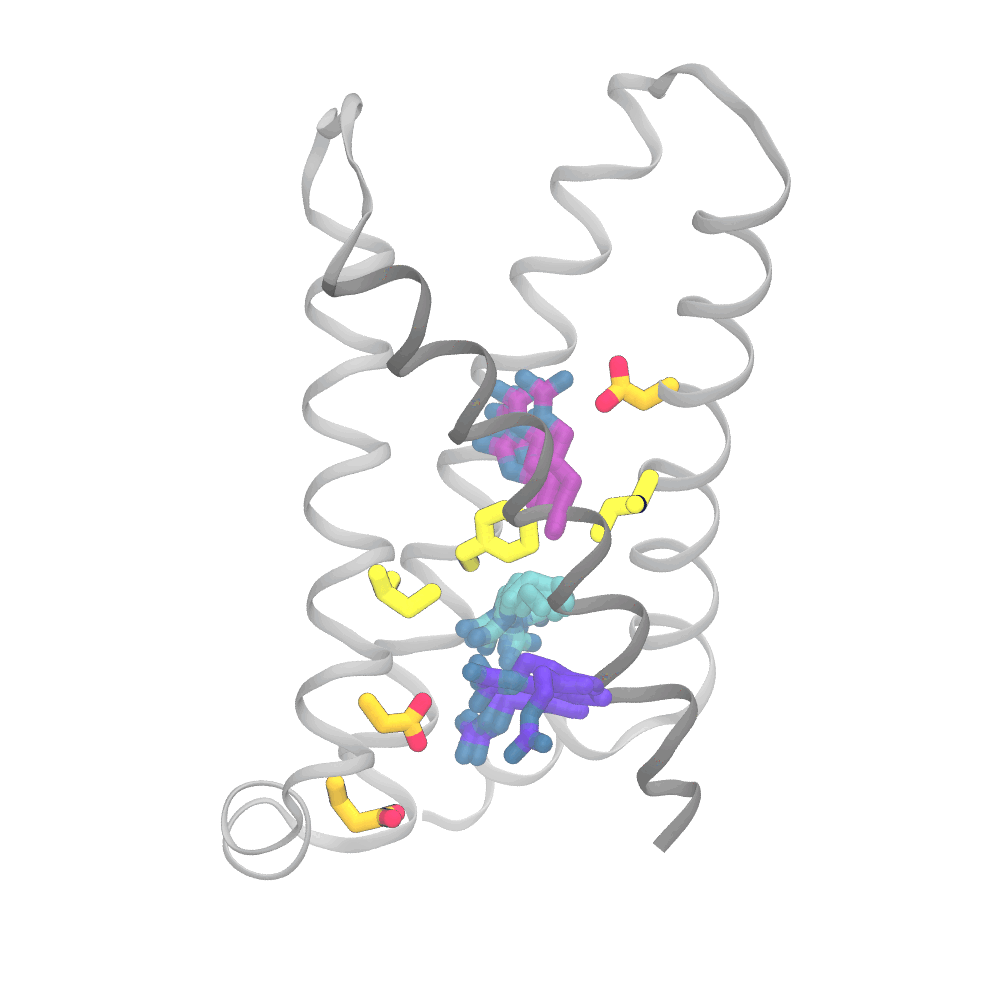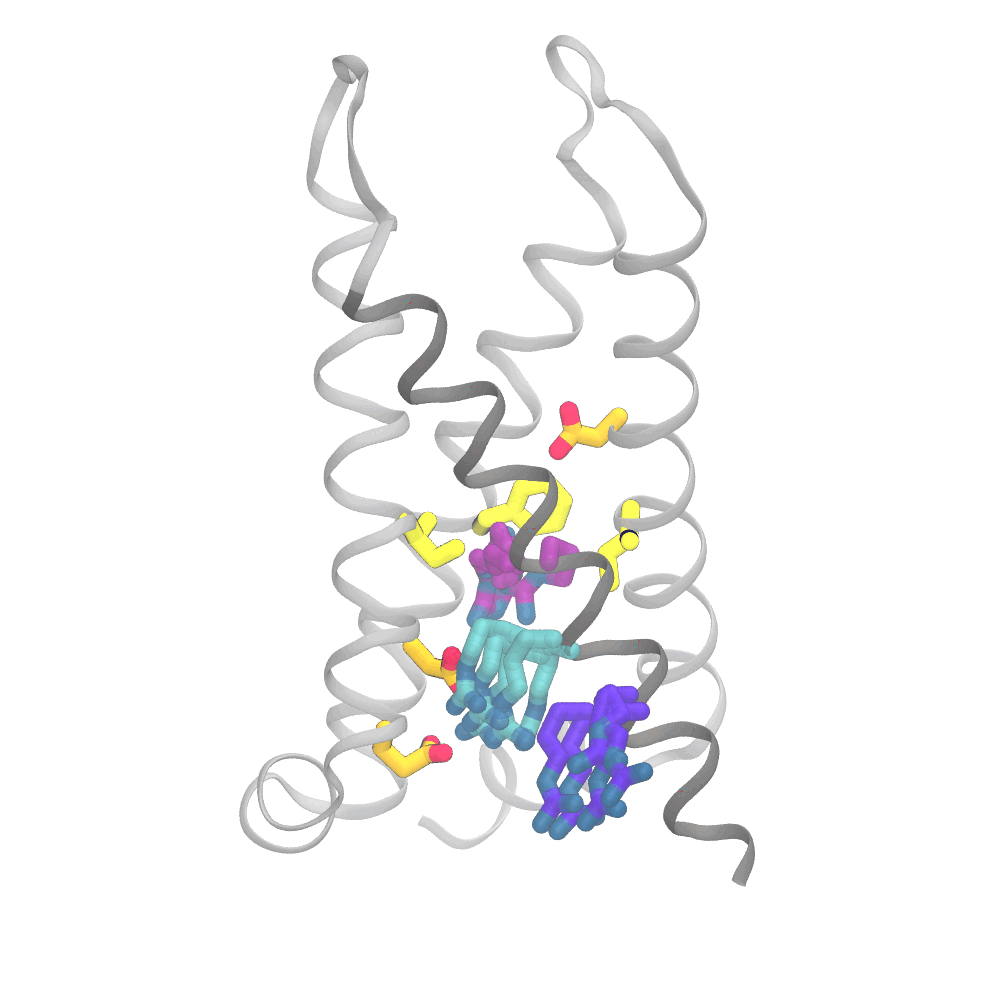
This animation shows the consensus simulation the scientists obtained from combining their runs on Anton 2. The important charged arginine residues (purple/cyan) on the S4 helix (grey) are essential for activating the protein. They do this by interacting with the negatively charged residues (orange) and the hydrophobic plug residues (yellow). Credit: Spencer Guo.
The simulations were eye opening. Rather than the “screw” part of the protein moving partly up and out of the socket in coordination with its rotation and the changes in interactions of the protein side chains, the three sets of movements were only loosely coupled. The outward movement, rotation, and changes in side-chain interactions could happen in varying order. Importantly, the work captured a number of states for the protein that hadn’t been directly detected by laboratory methods.
The team reported these results in the prestigious journal Nature Communications in February 2024. In particular, their paper suggests ways of validating their findings on real voltage-sensing phosphatase in the laboratory that could in turn spur better simulations. Such a game of ping-pong between lab and computer holds promise in sharpening scientists’ understanding of how these proteins work and how their behavior can be affected by drugs intended to reverse human diseases.
