
Adobe Stock #527516305.
Scientists leverage past MARC program to maintain competitive bioinformatics program at minority-serving institution
Lucina pectinata is a clam that lives in mud flats among levels of hydrogen sulfide high enough to harm most animals. Scientists from the University of Puerto Rico, leveraging training from the past MARC program and supercomputers at PSC, shed light on how the clam’s evolution resulted in hemoglobin proteins that prevent it from being poisoned by the gas while feeding a friendly bacteria. The work holds promise in developing improved detectors for toxins and a deeper understanding of how evolution helps organisms duplicate their proteins to create new chemical functions.
WHY IT’S IMPORTANT
Some of the most important lessons we’ve learned about how our bodies and the chemicals within them function come from studying animals that live in conditions that would kill us.
Consider Lucina pectinata, the thick lucine. A saltwater clam, it lives in tropical coastal mudflats with high levels of hydrogen sulfide — a toxic and corrosive gas that is the keynote in “rotten egg” smells. Scientists knew that the clam survived because it had evolved an alternate version of the hemoglobin protein that carries oxygen in animals’ bloodstream. This version has a mutation that allows it to bind hydrogen sulfide instead of oxygen. The “new” versions of the proteins help have mutations that protect the clam’s “normal” hemoglobins from the gas, while the sulfur-carrying protein transports it to a symbiotic microbe — one that lives within the clam with both species getting benefit — that detoxifies it.
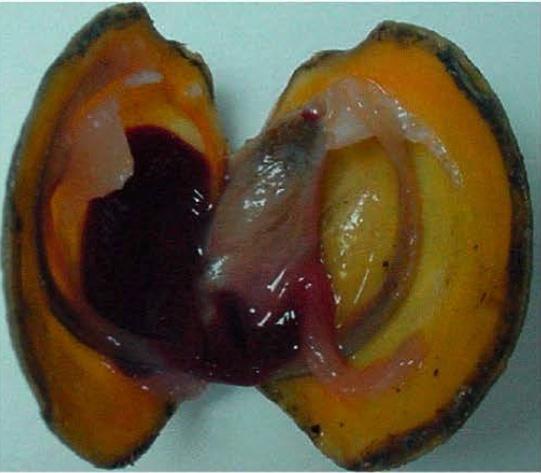
Lucina pectinata, the thick lucine saltwater clam. From Montes-Rodríguez, I.M.; Cadilla, C.L.; López-Garriga, J.; González-Méndez, R. Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins. Genes 2022, 13, 2041. https://doi.org/10.3390/genes13112041
“[Earlier] papers showed that, a long time ago, the clam evolved two hemoglobins — one to transport oxygen and one to transport hydrogen sulfide. Then, the oxygen-carrying version experienced a gene duplication … The structures are very similar, except for mutations that affect how they coordinate the heme center [that binds oxygen] and protect the [oxygen-carrying] hemoglobin from binding sulfur.” — Ricardo González-Méndez, UPR-Medical Sciences Campus
Some of the mystery about L. pectinata and its hemoglobins lifted when University of Puerto Rico–Mayagüez Campus graduate student and 2013 PSC summer intern Ingrid Montes-Rodríguez used BioU, a former PSC supercomputer dedicated to biological research, to unveil the clam’s genome and transcriptome — its DNA sequence and the sequence of RNAs that translate gene instructions into proteins. She earned her PhD with this work, done in part through the MARC program that the National Institutes of Health funded at PSC. MARC helped minority-serving institutions develop classes and programs to give their students the skills necessary to compete in the new, computerized science of bioinformatics.
Montes-Rodríguez’s findings were the gift that kept on giving. Once the genes had been revealed, scientists could mine it for more information on how the clam accomplished its evolutionary survival pivot. Carmen Cadilla of the University of Puerto Rico–Medical Sciences Campus; Juan López-Garriga, Montes-Rodriguez’s PhD advisor at UPR Mayagüez; and collaborator and MARC co-principal investigator Ricardo González-Méndez at the Medical Sciences Campus continued to work with her and her data to learn more.
HOW PSC HELPED
First envisioned by González-Méndez in the mid-1990s and building on PSC’s successful outreach efforts to Historically Black Colleges and Universities and at UPR, the Biomedical Initiative Group at PSC proposed a successful grant application in 2001. Led by PSC’s David Deerfield, Hugh Nicholas, and Alex Ropelewski, PSC’s MARC program proved fundamental in starting first-rate bioinformatics research at UPR Medical Sciences. Unfortunately, MARC funding ended in 2016. But González-Méndez and other faculty at UPR were determined to keep the bioinformatics work going. Happily, MARC had left the program with a deep bench of expertise in the field that enabled various faculty members to put together funding from other sources, including smaller grants from the NIH.
One of these efforts was the continued analysis of Montes-Rodríguez’s data. An investigation of the three types of hemoglobin coded by the clam’s DNA revealed an interesting evolutionary split from its near relatives without hydrogen-sulfide-binding hemoglobin.
“PSC’s resources were really important, because for a lot of students [at minority-serving institutions], all they had available was the Web and … [free] resources such as NCBI and GenBank … But then, when you got out of the really basic stuff, the work that needed to be done, such as being able to analyze genomes and transcriptomes, [required] in-depth phylogeny analyses [and large computing resources] … BioU and other PSC resources allowed these students to [do the] work …” — Ricardo González-Méndez, UPR–Medical Sciences Campus
Two of the L. pectinata hemoglobins, HBIILp and HBIIILp, carry oxygen to the clam’s tissues. But their amino acid sequences diverged more compared with the hemoglobins of “normal” clams than the hydrogen-sulfide-binding hemoglobin, HBILp. The oxygen-binding proteins’ more extensive mutations allow them to avoid being poisoned by hydrogen sulfide and to carry oxygen despite its presence. Meanwhile, a less-radical mutation in HBILp strengthens its reaction with hydrogen sulfide, making it more capable of sucking the poison from L. pectinata’s bloodstream. The scientists published their results in the journal Genes in November 2022.
The findings may have impacts in several biomedical fields. First, HBILp holds promise as a sensor for hydrogen sulfide in detecting toxic gases. Dr. López-Garriga has continued this work. The chemistry of how hemoglobin changed both to resist and embrace binding of the gas may also shed light on human genetic diseases involving oxygen binding, such as sickle-cell anemia and beta thalassemia. Finally, the larger question of exactly how a single protein becomes duplicated into different copies that then diverge to perform distinct functions lies at the core of how evolution works. The collaborators would like to build on their work, investigating all these important facets.
In more recent work with PSC, González-Méndez has worked with PSC’s Ropelewski to write a grant for a diversity program within the Brain Imaging Library. Ropelewski is principal investigator for this project, along with Alan Watson of the University of Pittsburgh. The funding will enable students to study high performance computing-enabled microscopic image analysis at PSC.
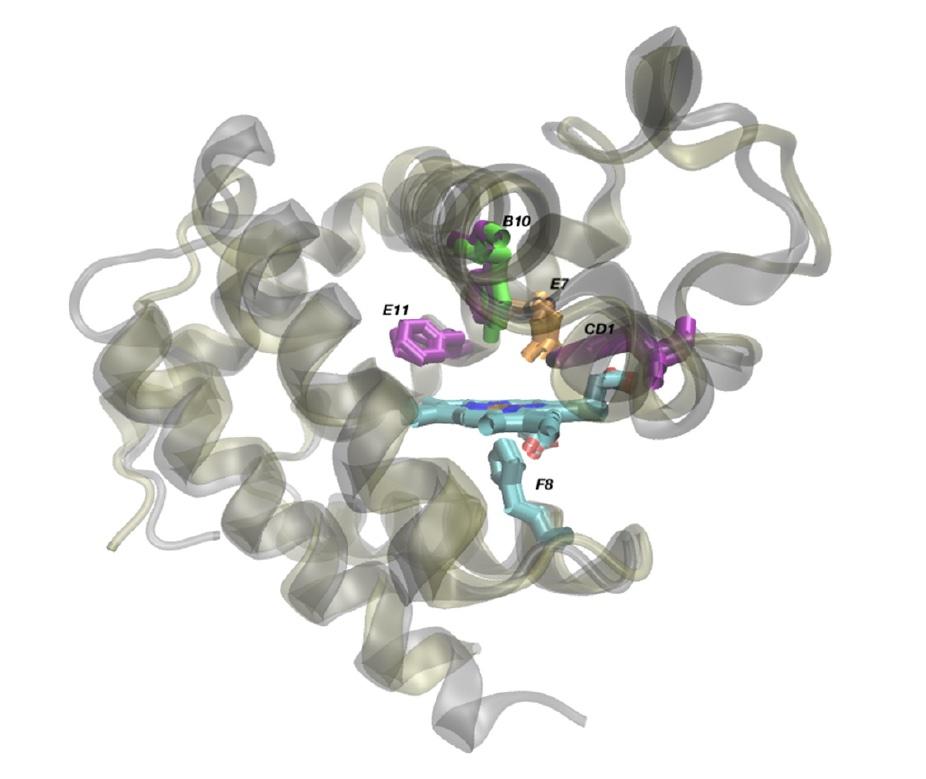
The L. pectinata HBILp and HBIILp proteins, superimposed to show the differences between them that make the former bind to hydrogen sulfide and the latter oxygen. The main difference comes down to position B10, where the HbILp structure has the amino acid phenylalanine and HbIILp has the amino acid tyrosine. From Montes-Rodríguez, I.M.; Cadilla, C.L.; López-Garriga, J.; González-Méndez, R. Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins. Genes 2022, 13, 2041. https://doi.org/10.3390/genes13112041
