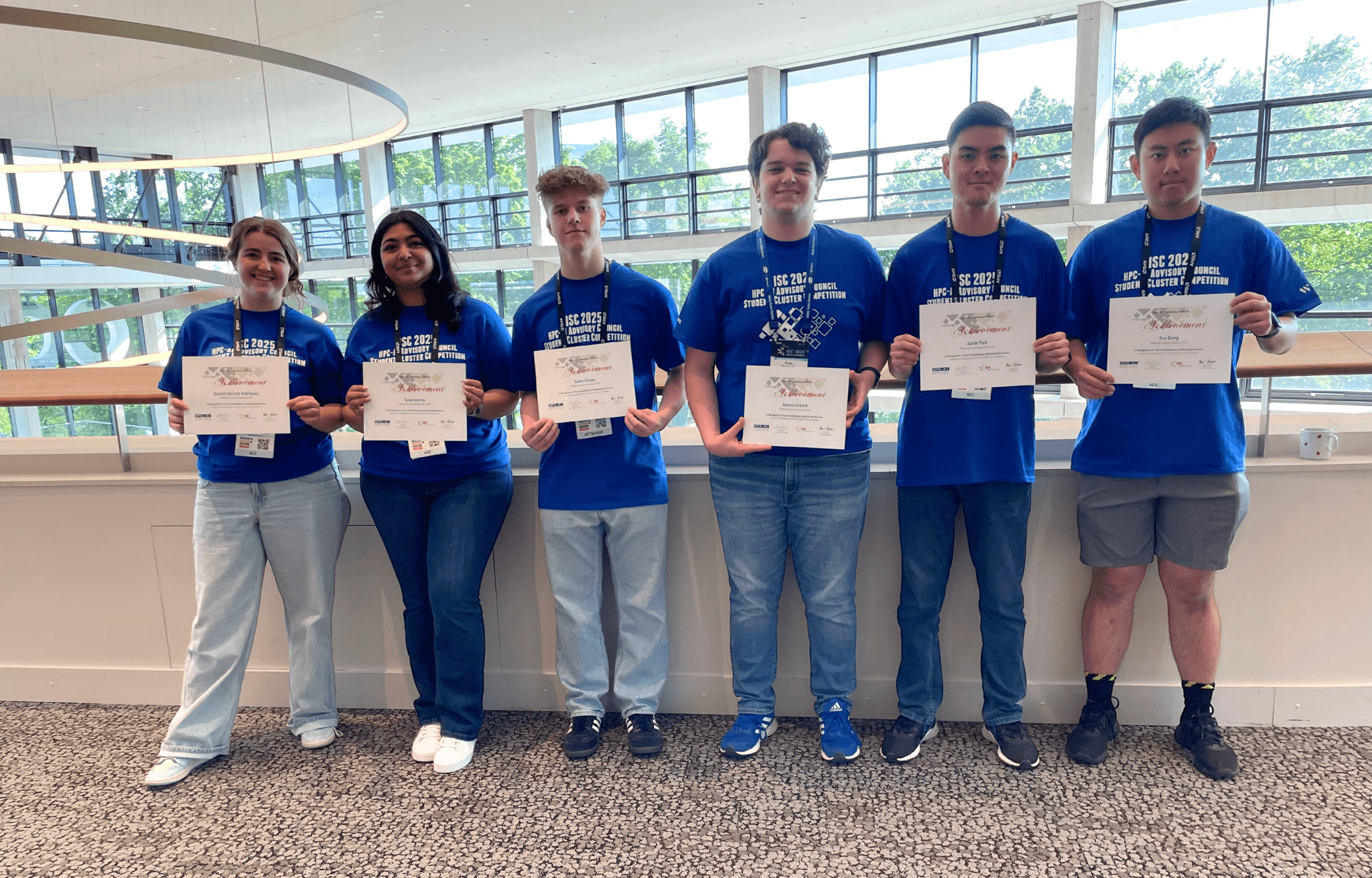
PSC Partners Again in ByteBoost Cybertraining Program
PSC joins with NSF-funded ACCESS partners Stony Brook University and Texas A&M University to host the ByteBoost Cybertraining Program.
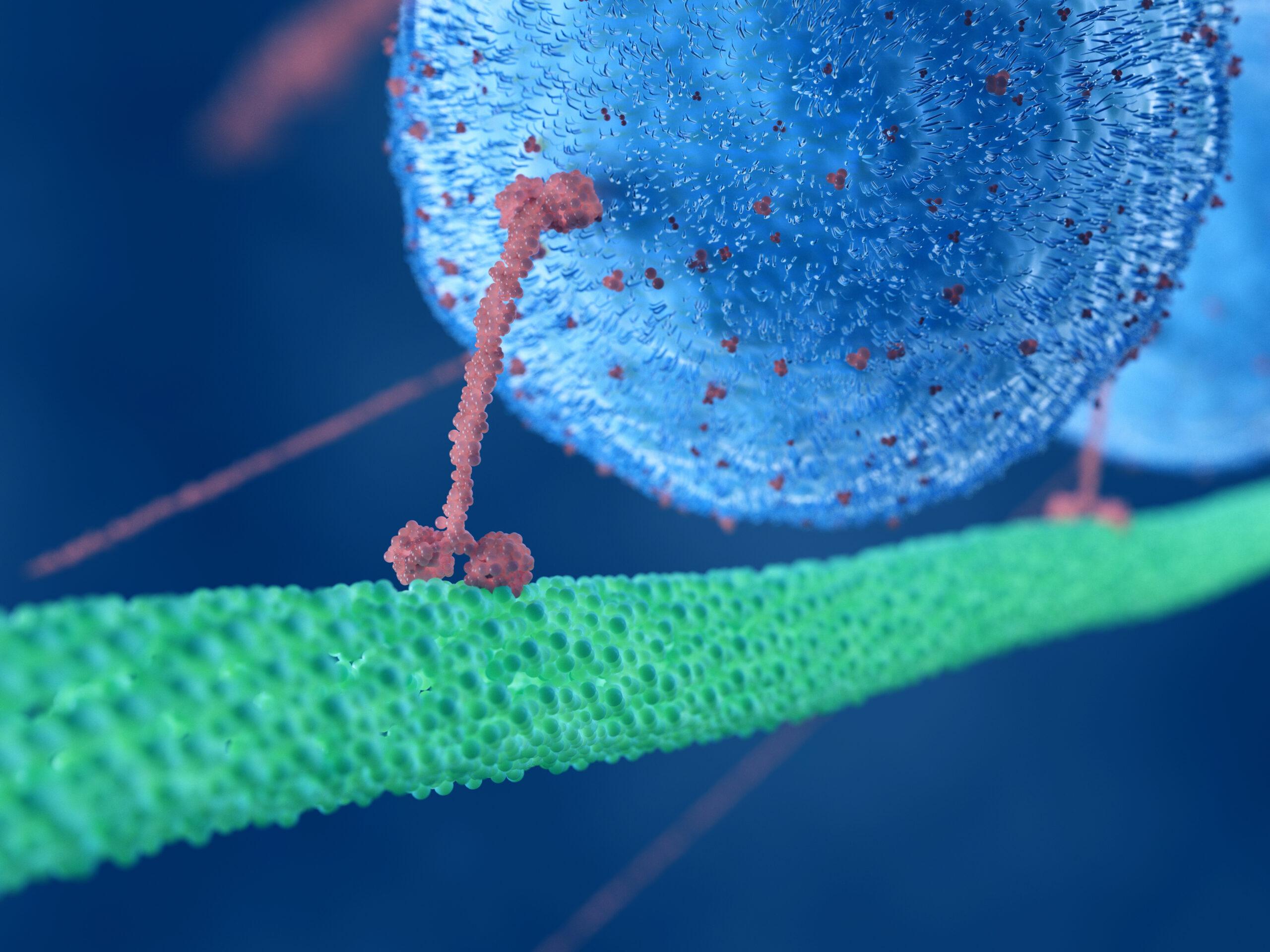
Anton Simulations Flag How Hepatitis Virus Enters Cell Nucleus
“Tucking In” of Viral Capsid Protein Allows Passage of Giant Structure through Nuclear Pore, Allowing Virus to Take Over
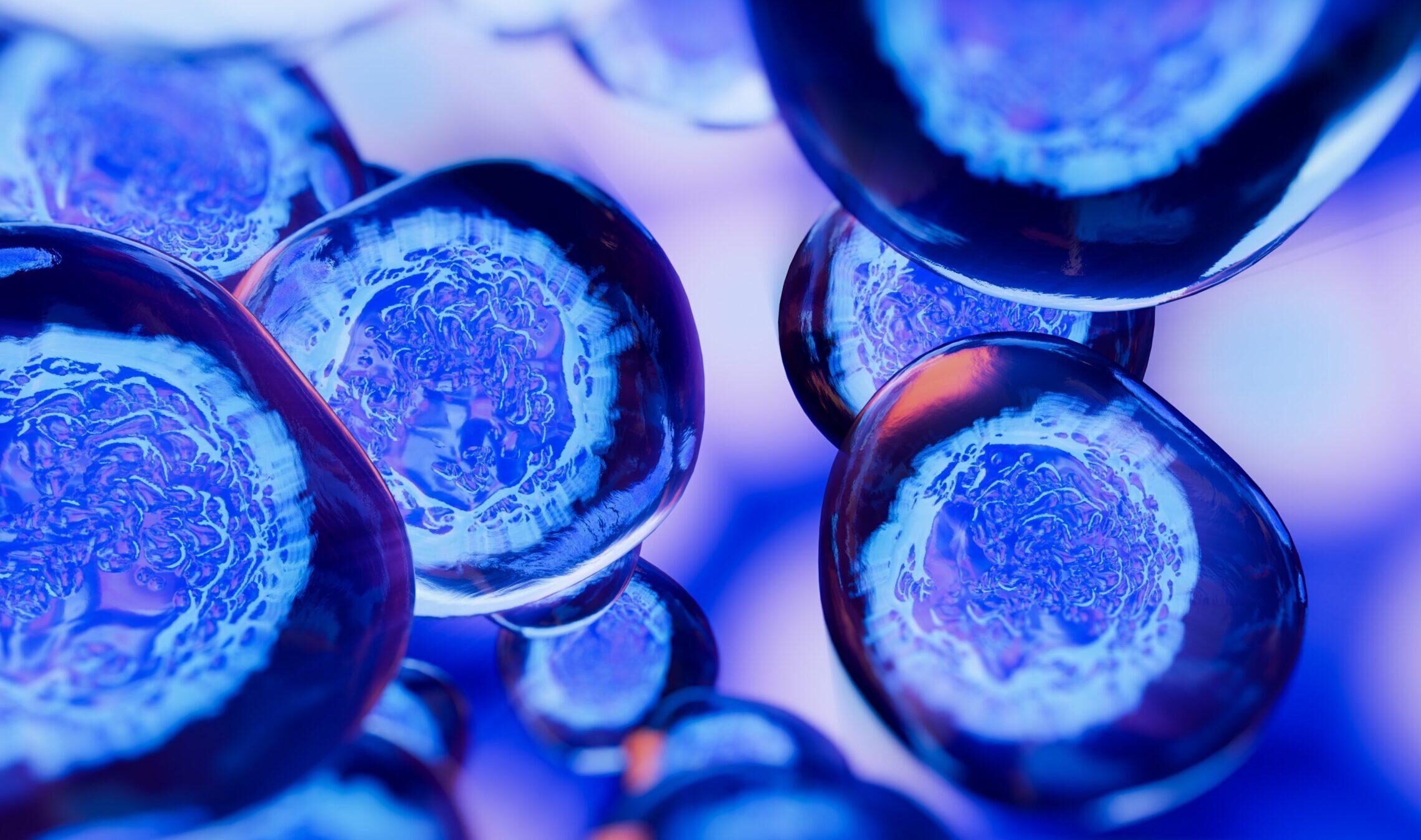
Bridges-2 and AI Analysis Identify DNA Sequences Carrying Risk of Childhood Sudden Death
Number and Type of Mutations Relate to Risk of Death at Earlier Age, Suggesting Focus for Future Detection and Prevention
Accelerate your research on Bridges-2,
our flagship supercomputer

Our featured projects
PSC maintains advanced infrastructure to support
computation-heavy research in areas such as: data analytics, machine learning, bimolecular simulation, AI and deep learning, and provides access to the national cyberinfrastructure community of resources.
Want to help further our research?
Support the next big discovery or inspire the next class of great thinkers with a gift to our center.

Recent News from PSC
PSC Partners Again in ByteBoost Cybertraining Program
PSC joins with NSF-funded ACCESS partners Stony Brook University and Texas A&M University to host the ByteBoost Cybertraining Program.
Anton Simulations Flag How Hepatitis Virus Enters Cell Nucleus
“Tucking In” of Viral Capsid Protein Allows Passage of Giant Structure through Nuclear Pore, Allowing Virus to Take Over
Bridges-2 and AI Analysis Identify DNA Sequences Carrying Risk of Childhood Sudden Death
Number and Type of Mutations Relate to Risk of Death at Earlier Age, Suggesting Focus for Future Detection and Prevention
Accelerate your research on
Bridges-2, our newest supercomputer

Our Featured Projects
PSC maintains advanced infrastructure to support computation-heavy research in areas in: data analytics, machine learning, bimolecular simulation, AI and deep learning, and provides access to the national cyberinfrastructure community of resources.
Want to help further our research?
Support the next big discovery or inspire the next class of great thinkers with a gift to our center.